Molecular
Beacons FAQ's Taq Man/Molecular
Beacons*/Fluorescent Probes Price List
Introduction
Molecular beacons are oligonucleotide probes that can report the presence of
specific nucleic acids in homogenous solutions (Tyagi S, Kramer FR. Molecular
beacons: probes that fluoresce upon hybridization, Nature Biotechnology 1996;
14: 303-308.) They are useful in situations where it is either not possible or
desirable to isolate the probe-target hybrids from an excess of the
hybridization probes, such as in real time monitoring of polymerase chain
reactions in sealed tubes or in detection of RNAs within living cells. Molecular
beacons are hairpin shaped molecules with an internally quenched fluorophore
whose fluorescence is restored when they bind to a target nucleic acid (Figure
1). They are designed in such a way that the loop portion of the molecule is a
probe sequence complementary to a target nucleic acid molecule. The stem is
formed by the annealing of complementary arm sequences on the ends of the probe
sequence. A fluorescent moiety is attached to the end of one arm and a quenching
moiety is attached to the end of the other arm. The stem keeps these two
moieties in close proximity to each other, causing the fluorescence of the
fluorophore to be quenched by energy transfer. Since the quencher moiety is a
non-fluorescent chromophore and emits the energy that it receives from the
fluorophore as heat, the probe is unable to fluoresce. When the probe encounters
a target molecule, it forms a hybrid that is longer and more stable than the
stem and its rigidity and length preclude the simultaneous existence of the stem
hybrid. Thus, the molecular beacon undergoes a spontaneous conformational
reorganization that forces the stem apart, and causes the fluorophore and the
quencher to move away from each other, leading to the restoration of
fluorescence.
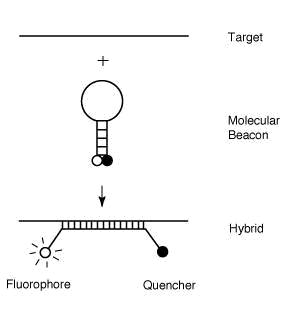
Figure 1. Operation of
molecular beacons. On their own, these molecules are non-fluorescent, because
the stem hybrid keeps the fluorophore close to the quencher. When the probe
sequence in the loop hybridizes to its target, forming a rigid double helix, a
conformational reorganization occurs that separates the quencher from the
fluorophore, restoring fluorescence.
In order to detect multiple targets in the same solution,
molecular beacons can be made in many different colors utilizing a broad range
of fluorophores (Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for
allele discrimination, Nature Biotechnology 1998; 16: 49-53.) DABCYL a
non-fluorescent chromophore, serves as the universal quencher for any
fluorophore in molecular beacons. Owing to their stem, the recognition of
targets by molecular beacons is so specific that single-nucleotide differences
can be readily detected.
Molecular
Beacon Example Sequence
Fluorophore at 5' end; 5'-GCGAGCTAGGAAACACCAAAGATGATATTTGCTCGC
-3'-DABCYL
The underlined sequence at the 5' and 3' ends identifies
the arm sequences that are complementary.
The length of the probe sequence (10-40 nt) is chosen in
such a way that the probe target hybrid is stable in the conditions of the
assay. The stem sequence (5-7 nt) is chosen to ensure that the two arms
hybridize to each other but not to the probe sequence. Folding of the designed
sequence with the help of a computer program can indicate whether the intended
stem-and-loop conformation will occur. The computer program can also predict the
melting temperature of the stem
Signal
to background ratio
1. Determine the fluorescence (Fbuffer) of 200 µl of molecular
beacon buffer solution using 491 nm as the excitation wavelength and 515 as the
emission wavelength. If the fluorophore is not fluorescein, chose wavelengths
that are optimal for the fluorophore in the molecular beacon.
2. Add 10 µl of 1 µM molecular beacon to this solution and record the new
level of fluorescence (Fclose).
3. Add a two-fold molar excess of the oligonucleotide target and monitor the
rise in fluorescence until it reaches a stable level (Fopen).
4. Calculate the signal to background ratio as (Fopen-Fbuffer)/(Fclose-Fbuffer).
Thermal
denaturation profiles
1. Prepare two tubes containing 50 µl of 200 nM molecular beacon dissolved in
3.5 mM MgCl2 and 10 mM Tris-HCl, pH 8.0 and add the oligonucleotide
target to one of the tubes at a final concentration of 400 nM.
2. Determine the fluorescence of each solution as a function of temperature
using a thermal cycler with the capacity to monitor fluorescence. Decrease the
temperature of these tubes from 80 °C to 10 °C in 1 °C steps, with each hold
lasting one min, while monitoring the fluorescence during each hold.
Real
time monitoring of polymerase chain reactions
Utilize molecular beacons that are complementary to a sequence in the middle of
the expected amplicon. The length of their arm sequences should be chosen so
that a stem is formed at the annealing temperature of the polymerase chain
reaction. The length of the loop sequence should be chosen so that the
probe-target hybrid is stable at the annealing temperature. Whether a molecular
beacon actually exhibits these designed features is determined by obtaining
thermal denaturation profiles. The molecular beacons with appropriate thermal
denaturation characteristics are included in each reaction at a concentration
similar to the concentration of the primers. During the denaturation step, the
molecular beacons assume a random coil configuration and fluoresce. As the
temperature is lowered to allow annealing of the primers, stem hybrids form
rapidly, preventing fluorescence. However, at the annealing temperature,
molecular beacons also bind to the amplicons and generate fluorescence. When the
temperature is raised to allow primer extension, the molecular beacons
dissociate from their targets and do not interfere with polymerization. A new
hybridization takes place in the annealing step of every cycle, and the
intensity of the resulting fluorescence indicates the amount of accumulated
amplicon.
Procedure
1. Set up six 50 µl reactions so that each contains a different number of
targets, 0.34 µM molecular beacon, 1 µM of each primer, 2.5 units of Amplitaq
Gold DNA polymerase (Perkin Elmer), 0.25 mM of each deoxyribonucleotide, 3.5 mM
MgCl2, 50 mM KCl, and 10 mM Tris-HCl, pH 8.0.
2. Program the thermal cycler to incubate the tubes at 95 °C for 10 min to
activate Amplitaq Gold DNA polymerase, followed by 40 cycles of 30 sec at 95 °C,
60 sec at 50 °C and 30 sec at 72 °C. Monitor fluorescence during the 50 °C
annealing steps.
Troubleshooting
Low signal-to-background ratio.
The assay medium may contain insufficient salt. There
should be at least 1 mM MgCl2 in the solution, in order to ensure
that the stem hybrid forms. The molecular beacon may fold into an alternate conformation that results in a
sub-population that is not quenched well. Change the stem sequence (and probe
sequence, if necessary) to eliminate that possibility.
Incomplete restoration of fluorescence at low
temperatures.
If the stem of a molecular beacon
is too strong, at low temperatures it may remain closed while the probe is bound
to the target. This may happen inadvertently if the probe sequence can
participate in the formation of a hairpin that results in a stem longer and
stronger than originally designed. Change the sequence at the edges of the probe
and the stem sequence to avoid this problem.
*Disclaimer of License Statement for Molecular Beacons Products
The 5' Nuclease detection assay and other homogeneous amplification methods used in connection with the Polymerase Chain Reaction ("PCR") Process are covered by patents owned by Roche Molecular Systems, Inc. and F. Hoffmann-La Roche Ltd. ("Roche"). Use of these methods requires a license. No license under these patents, which include but are not limited to United States Patent Nos. 5,210,015, 5,487,972, 5,804,375, and 5,994,076 to use the 5' Nuclease Assay or any Roche patented homogeneous amplification process is conveyed expressly or by implication to the purchaser by the purchase of any Gene Link, Inc. PCR-related product. Purchasers of these products must obtain a license to use the 5' Nuclease or Homogeneous PCR process before performing PCR. Further information on purchasing licenses to practice the PCR Process may be obtained by contacting the Licensing Specialist at (510) 814-2984, Roche Molecular Systems, Inc. 1145 Atlantic Avenue, Alameda, California, 94501
*PHRI Molecular Beacon License Agreement
"This product is sold under license from the Public Health Research Institute. It may be used under PHRI Patent Rights only for the purchaser's research and development activities".

|
|
|

